Intestinal Methanomassiliicoccales
This page evolves frequently... Do not hesitate to come back later (11/02/2018)
Les Methanomassiliicoccales, qu'est-ce ?
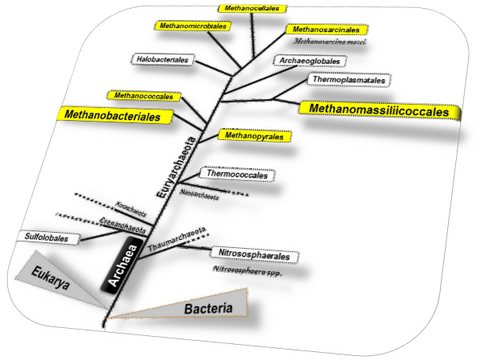
Methanomassiliicoccales are a taxonomic order within Archaea. A taxonomic order is composed of families, themselves composed of genera, species and strains. It is therefore a grouping of many species. Historically, the existence of Methanomassiliicoccales was considered around the 2000s, with the discovery of archaea related to Thermoplasmatales in various environments: the latter are aerobic microbes (manadtory need of O2) and thermo-acidophilic (life at pH 1 to 4, T ° around 45-65 ° C). In other words, the presence of such microbes in the rumen of many ruminant animals seemed incomprehensible, leading to the hypothesis of a possible methanogenic microbial group in this branch of life (see the work of Rustam Aminov in 2002 on the group of microorganisms RCC "Rumen Cluster C"), anaerobic and living close to neutral pH. This hypothesis ran up against various arguments, in particular:
- no other methanogen in this evolutionary branch at this time, making this hypothesis incompatible with the knowledge of the origin of methanogenesis and its evolution in the living.
- As shown in the figure, all methanogenic archaea (in yellow) were all from a central branch, making it unlikely that Methanomassiliicoccales are methanogenic organisms (in yellow on the left, close to Thermoplasmatales).
- no biochemical, metabolic and molecular arguments about methanogenesis
- no isolated or even cultured organisms to test these metabolic hypotheses.
2008-2010: unknown methanogenic in the human microbiota
A work published in 2017 by Guillaume Borrel in P. W. O'Toole 's laboratory (UCC-APC, Cork; Ireland) in collaboration with our group confirms the presence in humans of these 5 different species of Methanomassiliicoccales. It is the result of an in-depth analysis of sequence databases, the bibliography and especially more than one hundred metagenomes (ELDERMET study). It is possible, however, that other species are present. It is now especially proven that these species do not share the same genetic and metabolic potential, and that a large genomic variability seems to exist within the same species (Borrel et al, 2017).
One isolate, several genomes: first clues of the originality of these organisms
A first species of Methanomassiliicoccales was isolated in 2012 by the combined efforts of Didier Raoult's laboratory and that of Bernard Ollivier (Dridi et al, 2012). The name of this species, attributed by the discoverers, is Methanomasiliicoccus luminyensis. It automatically induces the name given to the family (higher taxonomical rank) and to the order (taxonomic rank above) if the species can not be attached to a taxonomic rank already described: thus was born the family of Methanomassiliicoccaceae and the order of Methanomassiliicoccales. Didier Raoult's group has also determined the almost complete genome of this isolate (Aurore Gorlas et al, 2012), which is a large one (about 2.7 Mbp). Its annotation has been done automatically / non-expertly by GenBank (determination of the genes, the supposed functions for which they code, ...). One month after this genome has been announced, we also publish the genome of another member of the order Methanomassiliicoccales, belonging to a different genus (Ca. Methanomethylophilus alvus, Borrel et al, 2012) next of another species likely belonging to the same genus Methanomassiliicoccus (Ca. Methanomassiliicoccus intestinalis, Borrel et al, 2013). In collaboration with Simonetta Gribaldo (Institut Pasteur Paris), we gave the finale proofs (based on phylogenomics), that Methanomassiliicoccales were another methanogenic order on a unexpected branch of the tree of life (Borrel et al, 2013).
An original methanogenesis
With the first elements of knowledge of known Methanomassiliicoccales, their methanogenesis was questioned:
- the isolate M. luminyensis (2012) shows an original methanogenesis made from methanol (as for methylotrophic methanogens) but requiring H2 as an electron donor (Dridi et al, 2012). This exception is not unique, especially for an inhabitant of the intestinal microbiota, because the same type of methanogenesis is carried out by Methanosphaera stadtmanae, a member of Methanobacteriales found more particularly in digestive microbiota (see here).
- Genome analysis of Ca. Methanomethylophilus alvus (Borrel et al, 2012) and others quickly established that the genes for a so-called "hydrogenotrophic" methanogenesis were totally absent (Borrel et al, 2013); In addition, a first comparative analysis of the available genomes (3 in 2013-2014) showed the absence of certain elements allowing the electron / proton transfers and the energy recovery once the methane molecule has been synthesized (Borrel et al. al, 2014). Various hypotheses have therefore been published (Borrel et al, 2014, Lang et al, AEM, 2015). In particular, the work of Uwe Deppenmeier group in Germany indicates a new mechanism, supported by experimental data for Methanomassiliicoccus luminyensis (Kröninger et al, FEBS Journal, 2016).
- From the known genome of Ca. Methanomethylophilus alvus (Borrel et al, 2012), we hypothesized that this species could use other simple methylated compounds than methanol. In particular, the genes allowing the use of methylamines (mono-, di- and trimethylamine, MMA, DMA and TMA) or dimethyl sulfide (DMS) are present, as is the case with Methanosarcinales of the family of Methanosarcinaceae, those which achieve a "pure" methylotrophic methanogenesis.
- In addition, the metabolism of methylamines to methane requires very original enzymes: they are methyltransferases (Mt) having a rare amino acid, pyrrolysine or Pyl in their active site. The acquisition of this amino acid is done by a modified genetic code, making pyrrolysine the 22nd protein-forming amino acid, while living beings usually have 20 protein-forming amino acids, encoded by an almost universal genetic code. The case of Methanomassiliicoccales seems very original (Borrel et al, 2014b), explaining that we are devoting a part of our work to it.
Pyrrolysine in Methanomassiliicoccales.
- We have provided experimental evidence that this methanogenesis could be achieved from methylamines from the initially isolated strain (Brugère et al., 2014), as well as from a member of Methanomethylophilus alvus species (Borrel et al, 2017). However, some Methanomassiliicoccales do not use all methylamines (see, for example,in termite, with a strain that does not use di- and trimethylamine, Lang et al, 2014). This is also apparently true in the digestive tract of humans: some Methanomassiliicoccales lack the genetic ability to use di- and trimethylamine, or even the entirety of simple methylamines (Borrel et al, 2017). Moreover, the latter do not have a pyrrolysine system.
This natural activity of certain strains is at the origin of the concept of archaebiotics.
Find out more about archaebiotics
Variations in elderly people according to their health?
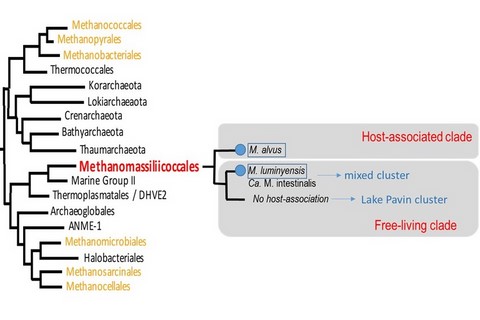
Surprisingly, Methanomassiliicoccales seem to be divided into two major lineages. One is apparently composed of species found only in digestive environments (insects, ruminants, man, ...) while a second lineage is mainly composed of species found in very diverse environments such as freshwater lake sediments, seep-sea oceans or in peatlands, etc. (Borrel et al, 2013, confirmed by Söllinger et al, 2016 or Borrel et al, 2017). The first compound lineage is said to be "host-associated", further names "host-associated clade". The second is named in opposition "Free-living clade", group that does not seem to live with a host. There are however 2 currently known exceptions of members of the free-living clade living in a host:
- Methanomassiliicoccus luminyensis belongs to this clade, while it has been isolated from human stool, and has not been found for the moment in other environments. However, it is difficult to say that it is frequently present in humans: apart from its isolation, it has not been found safely in humans, the tools used to confuse with a second member of this clade that we know present in humans.
- This second member is the candidate species Ca. Methanomassiliicoccus intestinalis. Surprisingly, he seems to be associated with unhealthy elderlies where it seems to surpass / replace the "Host-associated" clade members. What are the consequences and origins? In any case, this species questions about a possible role of pathobiont within archeal species.